Elucidating the Binding Mode between Heparin and Inflammatory Cytokines by Molecular Modeling - Yu - 2021 - ChemistryOpen - Wiley Online Library
Fine-tuning of the substrate binding mode to enhance the catalytic efficiency of an ortho-haloacetophenone-specific carbonyl reductase - Catalysis Science & Technology (RSC Publishing)
A computationally designed binding mode flip leads to a novel class of potent tri-vector cyclophilin inhibitors - Chemical Science (RSC Publishing)
Prediction of kinase inhibitors binding modes with machine learning and reduced descriptor sets | Scientific Reports
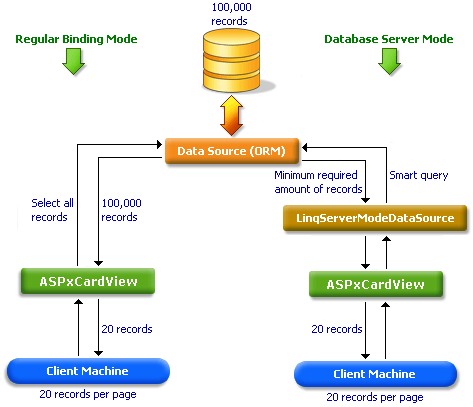
Bind Card View to Large Data (Database Server Mode) | ASP.NET Web Forms Controls | DevExpress Documentation

BSM00020: Difference of binding modes among three ligands to a receptor mSin3B corresponding to their inhibitory activities - BSM-Arc

The crystal structure of the second Z-DNA binding domain of human DAI (ZBP1) in complex with Z-DNA reveals an unusual binding mode to Z-DNA | PNAS
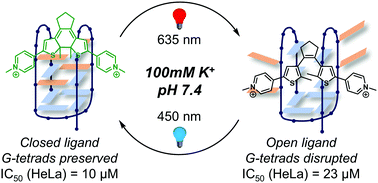
Visible-light photoswitching of ligand binding mode suggests G-quadruplex DNA as a target for photopharmacology - Chemical Communications (RSC Publishing)

Prediction of ligand binding mode among multiple cross-docking poses by molecular dynamics simulations | SpringerLink
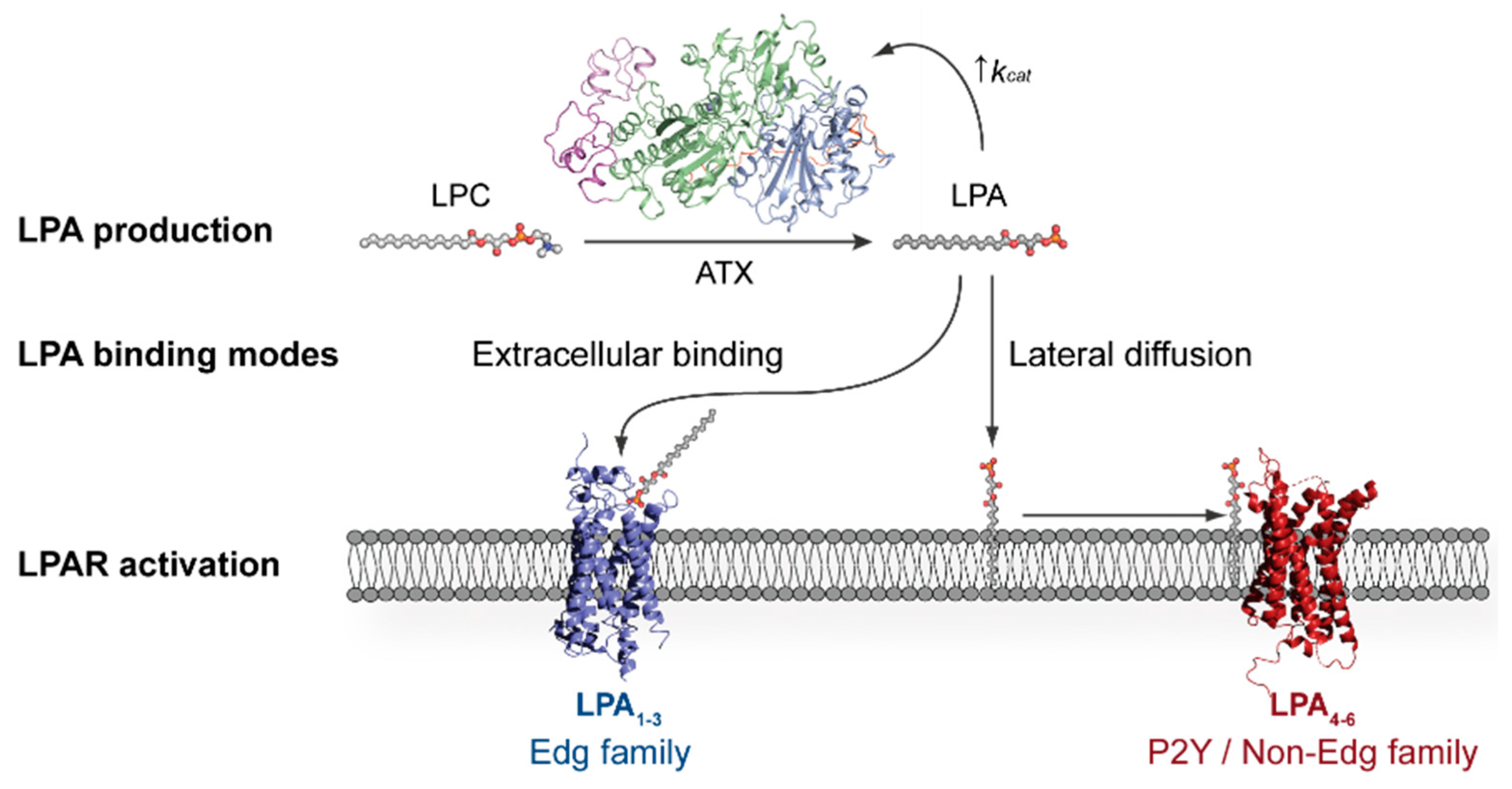
Cancers | Free Full-Text | The Structural Binding Mode of the Four Autotaxin Inhibitor Types that Differentially Affect Catalytic and Non-Catalytic Functions | HTML

Binding Modes of Ligands Using Enhanced Sampling (BLUES): Rapid Decorrelation of Ligand Binding Modes via Nonequilibrium Candidate Monte Carlo | The Journal of Physical Chemistry B
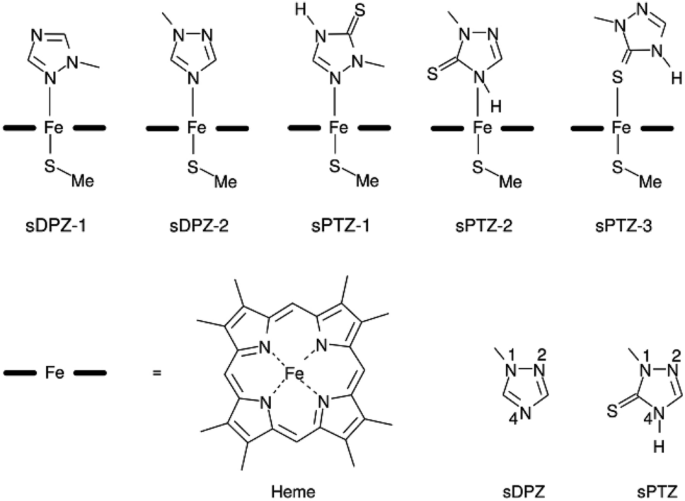
A binding mode hypothesis for prothioconazole binding to CYP51 derived from first principles quantum chemistry | SpringerLink
Computer-guided binding mode identification and affinity improvement of an LRR protein binder without structure determination | PLOS Computational Biology

Structural Basis of Constitutive Activity and a Unique Nucleotide Binding Mode of Human Pim-1 Kinase* - Journal of Biological Chemistry

Binding mode and chemical interactions of lead molecules with residues... | Download Scientific Diagram
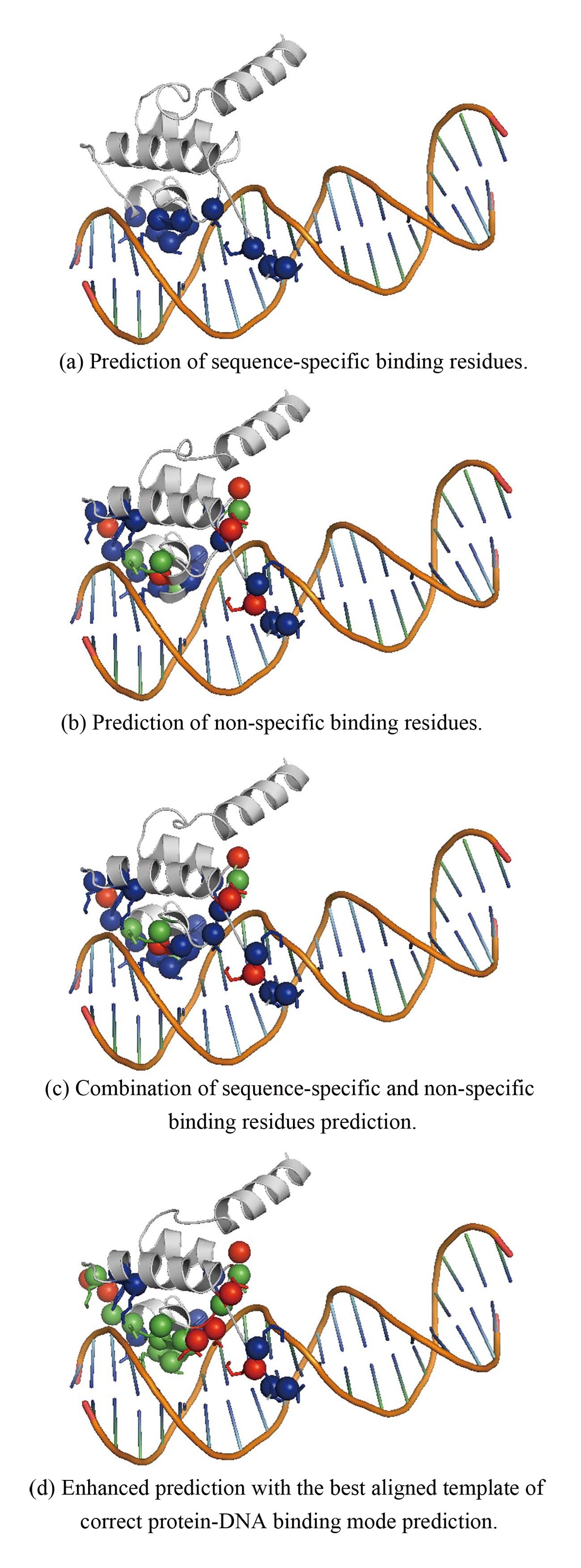
DNA-binding residues and binding mode prediction with binding-mechanism concerned models | BMC Genomics | Full Text


![PDF] A novel hypothesis for the binding mode of HERG channel blockers. | Semantic Scholar PDF] A novel hypothesis for the binding mode of HERG channel blockers. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/a5ba8ca2f388baea52f347b8f99ab27368584f24/2-Figure1-1.png)